Researchers from TU Delft and LUMC Leiden have developed a technique that can identify rare cell types within a sample of millions of cells. Immunologists hope it will provide a better understanding of our immune system.
CyTOF set-up. (Photo: Ecole Polytechnique Féderale de Lausanne)
Since 2013, immunologists at Leiden LUMC have been using a new research tool. An orange machine in the laboratory, called CyTOF, analyses 2,000 cells per second at the atomic level. In the lab, different cells can be marked with metal ions that bind onto specific antibodies. In the CyTOF machine, the cells are evaporated, and the metal ions are then analysed by a mass spectrometer to establish what kind of cell just flew past.Hundreds of cell types can now be distinguished. The costly cytometry by time of flight (CyTOF) analyser is also used by the Dutch Cancer Institute (NKI), the Sanquin blood bank, and the Erasmus MC. Researchers expect that this cell-specific precision of CyTOF will bring about advancements in immunology, parasitology and autoimmune diseases like rheumatism and Crohn’s disease.
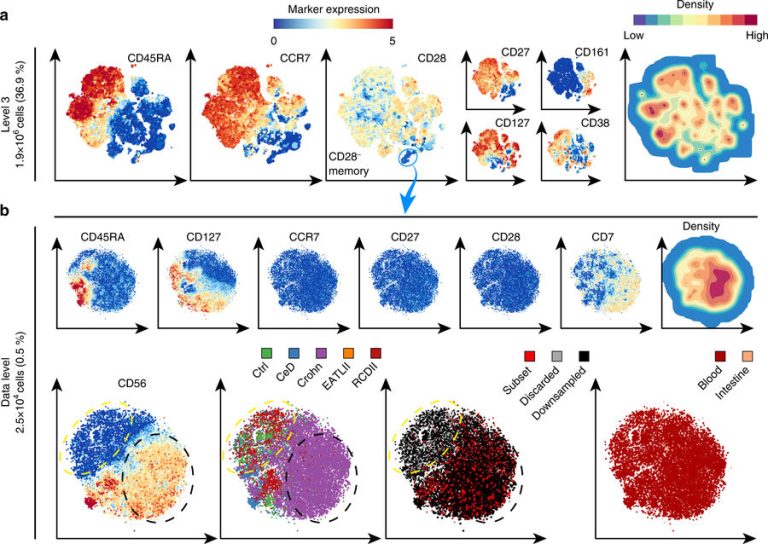
The CyTOF produces information of millions of cells with dozens of properties. In data terms, this is a multidimensional data cloud that needs to be analysed to identify new cell types. However, the size of the data makes it hard to see the wood for the trees. Dr. Anna Vilanova (EWI Faculty) and Professor Boudewijn Lelieveldt (LUMC and TU Delft) took the initiative for an improved data processing tool. LUMC immunologist Vincent van Unen explains: “Existing methods of analysing CyTOF data gave either a global overview of all the cells, or detailed information of an arbitrary part. But the most interesting cell types in a tissue sample, the ones that are linked to sickness or health, are often quite rare and easy to miss when you zoom in on a small part of the analysis.”
 Screen shot of the HNSE software (Photo: TU Delft)
Screen shot of the HNSE software (Photo: TU Delft)TU Delft researchers, led by Dr. Anna Vilanova from Professor Elmar Eiseman’s group computer graphics and visualisation at the EWI Faculty, developed a new analysis technique that allows immunologists to interactively zoom in onto interesting cell types without losing the oversight. PhD candidate Nicola Pezzotti, who developed the basic hierarchical analysis methodology (Hierarchic Stochastic Neighbour Embedding-HSNE), compares the technique to Google Earth: “You can start with the entire Earth and then zoom in on the street and house where you live. In our overview, we leave out the details, but we use the details to compute the landmarks.”
These ‘landmarks’ are a crucial part of the analysis. These are areas of cells that are alike, such as specific cells from the immune system. Computer scientist Dr Thomas Höllt, who developed the visualization strategy and works both at LUMC and TU Delft, explains: “By zooming in, you can discover rare cell types that are specific to certain diseases like Crohn’s disease, a chronic bowel syndrome. The presence of these rare cells provides clues to better understand the disease, diagnosis and targeted treatment.”
The new analysis technique, available in the software Cytosplore+HNSE (Hierarchic Stochastic Neighbour Embedding), was published in Nature Communications on 23 November 2017. The software is available at the Cytosplore website.
Vincent van Unen, Thomas Höllt and Nicola Pezzotti contributed equally to this work under supervision of Frits Koning (LUMC), Anna Vilanova and Boudewijn P.F. Lelieveldt.
The researchers made a demonstration video that shows the interactive exploration of their data.
Do you have a question or comment about this article?
j.w.wassink@tudelft.nl


Comments are closed.