An extra chromosome in yeast functions as a ‘playground’ for researchers. It allows them to design DNA and have a yeast cell stitch it together into a cellular factory.
(Illustration: Sija van den Beukel)
It’s hard to overestimate the merits of yeast. This unicellular organism has provided humanity with beer, bread and wine since antiquity. At present, yeast also produces biofuel and insulin. In the bio-based 21st century, the role of yeast (Saccharomyces cerevisiae) may well expand beyond the production of food, biofuels, pharmaceuticals and more. In fact, it now seems that any organic compound may be produced in or by yeast as long as it can be synthesised by enzymes and it’s not too toxic to the cell. “We’re trying to push the limits,” says senior researcher Professor Pascale Daran-Lapujade from the Applied Sciences Faculty. “We want to see how far we can go, what we can do.”
Daran-Lapjuda explains what has been achieved thusfar: “We have shown that a synthetic chromosome, designed on a computer, can be assembled from many DNA parts into something that is stably replicated, produces the number of copies expected, is transmitted from mother to daughter cell and is functional. These are the main findings of our work.”
As a proof of principle, PhD student Eline Postma is currently assembling extra chromosomes dedicated to producing anthocyanins, a class of compounds with a wide range of applications ranging from medical to food & feed utilisations. She identified the 40 genes in the pathway from sugar (input) to the end product, assembled them on an extra synthetic chromosome in the yeast nucleus and, hey presto, it churns out the product.
Seamless stitching
Let’s take a step back to see things in perspective. The artificial extra chromosome that the researchers worked with is about 100 kbp in size and about 100,000 base pairs long. Remember that the bases A, T, C and G are the letters of the genetic language, coding for the production of enzymes for example.
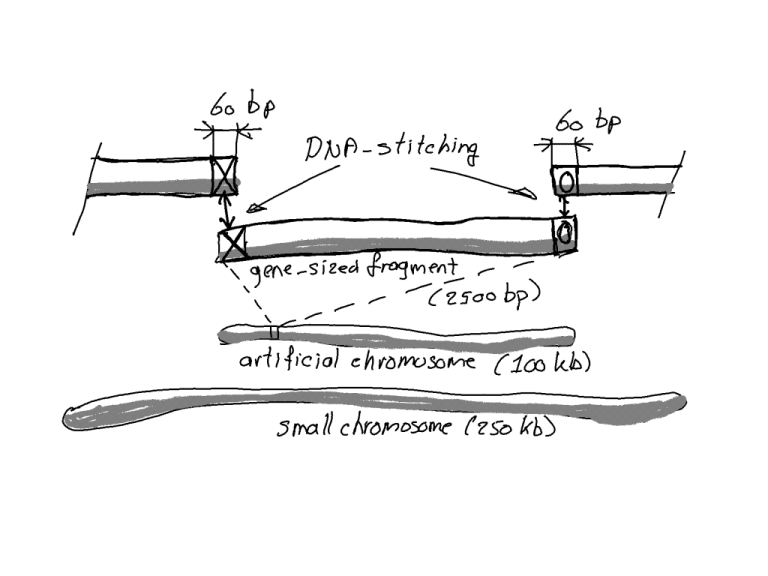

Basepairs in comparison. (Illustration: Jos Wassink)
Compare 100 kbp with the typical gene size of 2.5 kbp and with the smallest natural chromosome in yeast which is about 250 kbp. In short: at 100 kbp, the artificial chromosome is small, yet in the same size range as natural chromosomes. The researchers believe that it will also be possible to produce longer chromosomes.
The artificial chromosome is synthesised in overlapping fragments of about 2,500 bases, which are stitched together into a circular chromosome.
“Yeast is really a genome foundry,” says Daran-Lapujade. “It can assemble chromosomes of many different species. It is more efficient than any in-vitro (in the lab, eds.) method that we know. Baker’s yeast stitches DNA together so faithfully with hardly any mistakes. That’s really amazing.”
The ‘stitching’ process selects identical DNA sequences (in this case 60 bases long) and a cellular mechanism, essential for yeast to mend damaged DNA and chromosomes, then fuses these sequences together.
Two tracks
The possible applications thus seem to bifurcate. One application of the synthetic yeast chromosome is a platform on which researchers can assemble all the genetic machinery they need for the production of a certain compound. One of the main advantages of putting these genes together on an extra chromosome is the absence of interference with the rest of the genome.
Another application is the production of artificial chromosomes from scratch for other species such as bacteria. Or for the artificial cell that the Basyc consortium is developing.
“We regard it as a research tool,” says Daran-Lapujade, “We work in an industrial context with industrial microbes, but now we’re doing a fundamental study exploring the potential of modular chromosome assembly in yeast.”
- Eline Postma, Pascale Daran-Lapujade, A supernumerary designer chromosome for modular in vivo pathway assembly in Saccharomyces cerevisiae, Nucleic Acids Research, 11 January 2021.
Do you have a question or comment about this article?
j.w.wassink@tudelft.nl


Comments are closed.