Genes that didn’t seem important in the development of cancer happen to play a role after all. Delft researchers have discovered new areas in the genome where malicious mutations can occur.
Understanding of the three dimensional organization of the genome is paramount in finding genes that, when mutated, cause cancer. That is in a nutshell the conclusion of bioinformatics researcher Dr. Jeroen de Ridder and PhD candidate Sepideh Babaei of the section pattern recognition and bioinformatics (EEMCS faculty).
Together with colleagues from the Netherlands Cancer Insitute they developed a new tool to track down areas in the genome where the chances of the occurrence of malicious mutations are high. Their findings were published last week in Nature Communications.
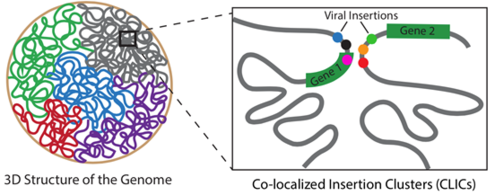
The researchers used a technique called insertional mutagenesis whereby viruses are used to cause mutations. Because many viruses insert their own genome into the genome of their host cells in order to replicate, mutagenesis caused by viral infections is quite common. The researchers found that the insertions tended to cluster in 3D hotspots and that genes that were far apart from each other linearly speaking (that is if one would unwind the tightly intertwined DNA) affected each other resulting is extra deregulation of cancer genes.
The findings shed new light on the repertoire of targets obtained from insertional mutagenesis screening and underline the importance of considering the genome as a 3D structure when studying effects of genomic perturbations.
S.Babaei et. al., 3D hotspots of recurrent retroviral insertions reveal long-range interactions with cancer genes. Nature Communications. February 27, 2015


Comments are closed.